In the week 6 and 7, I coded BayesianGaussianMixture for the full covariance type.
Now it can run smoothly on synthetic data and old-faithful data. Take a peek on the demo.
from sklearn.mixture.bayesianmixture import BayesianGaussianMixture as BGM
bgm = BGM(n_init=1, n_iter=100, n_components=7, verbose=2, init_params='random',
precision_type='full')
bgm.fit(X)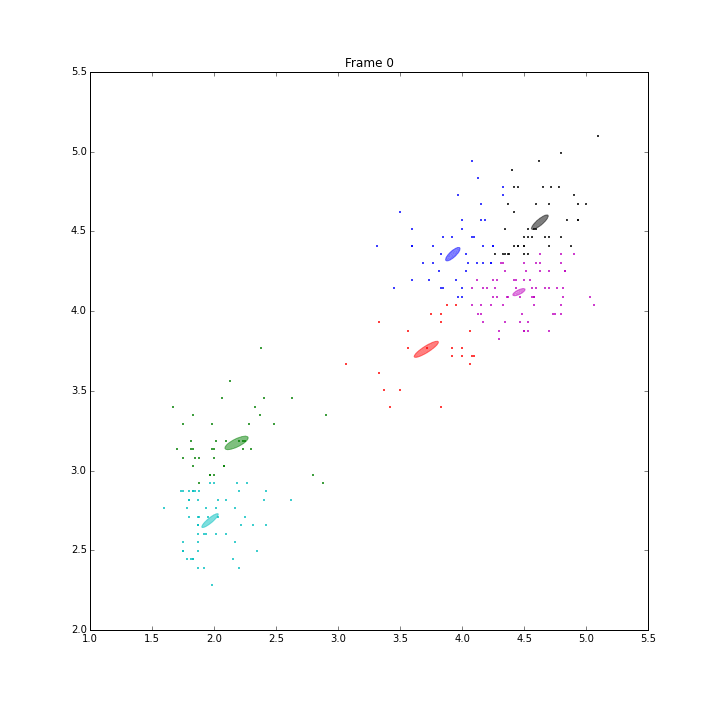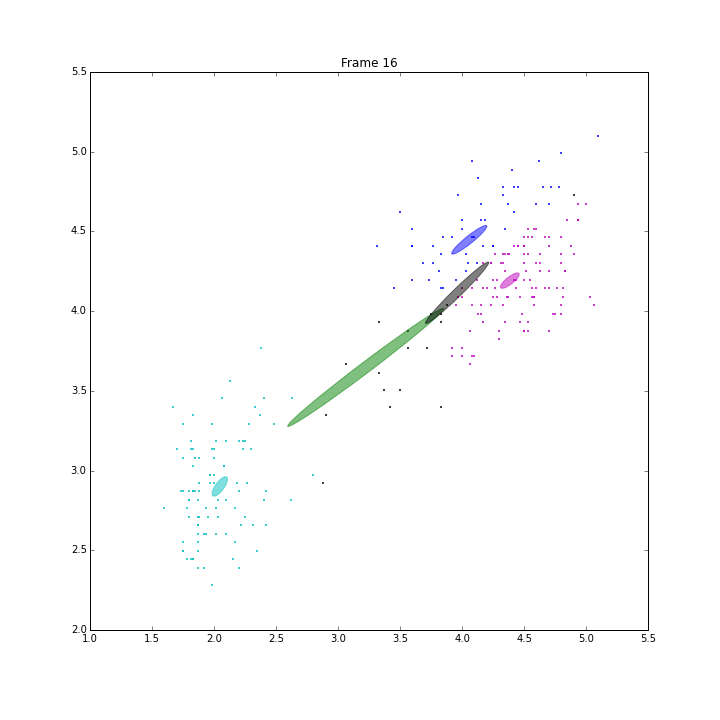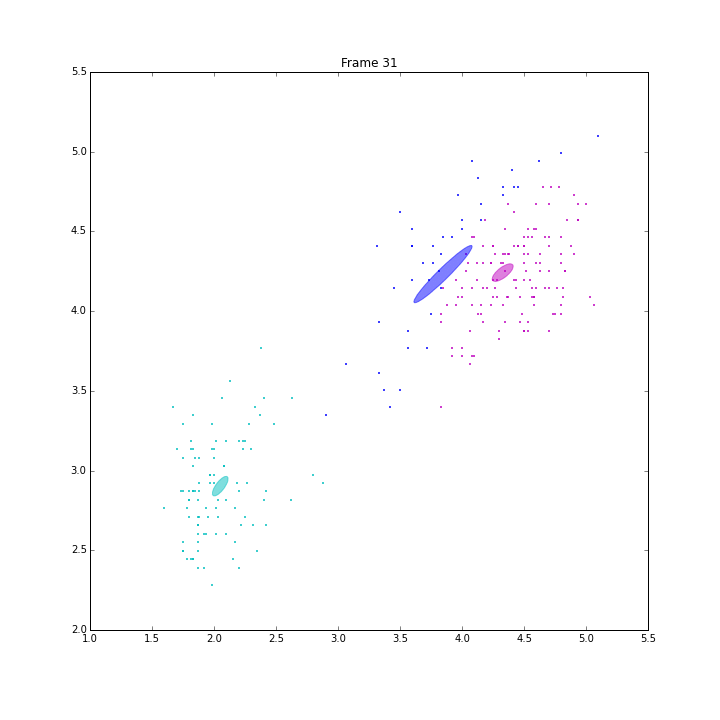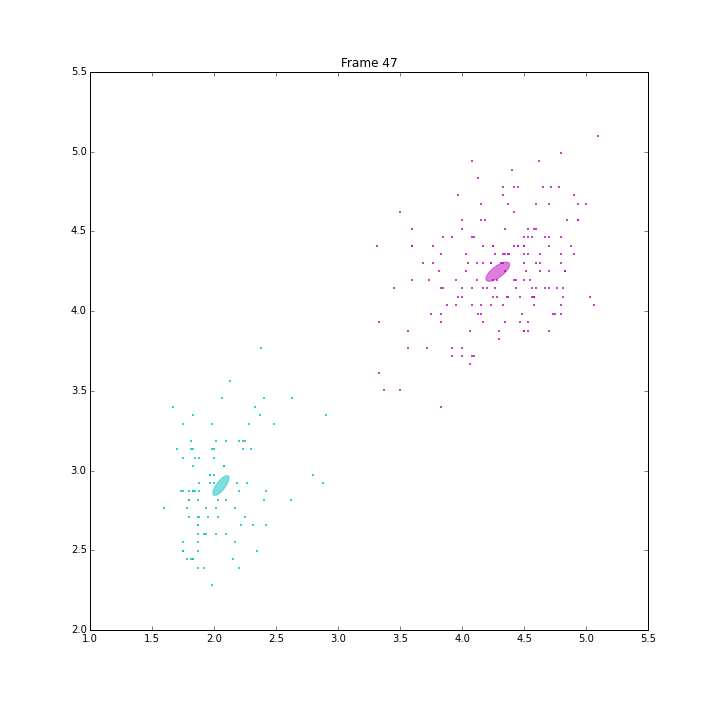
The demo is to repeat the experiment of PRML, page 480, Figure 10.6. VB on BGMM has shown its capability of inferring the number of components automatically. It has converged in 47 iterations.
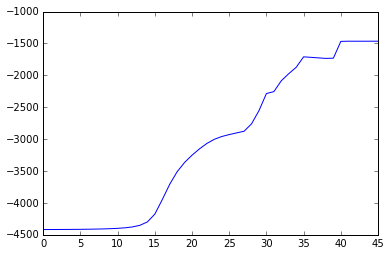
The ELBO looks a little weired. It is not always going up. When some clusters disappear, ELBO goes down a little bit, then go up straight. I think it is because the estimation of the parameters is ill-posed when these clusters have data samples less than the number of features.
The BayesianGaussianMixture has much more parameters than GaussianMixture, there are six parameters per each components.
I feel it is not easy to control the so many functions and parameters. The initial design of BaseMixture is also not so good.
I took a look at bnpy which is a more complicated implementation of VB on various mixture
models. Though I don’t need to go such complicated implementation, but the decoupling of observation model, i.e. $X$, $\mu$, $\Lambda$,
and mixture mode, i.e. $Z$, $\pi$ is quite nice. So I tried to use Mixin class to represent these two models. I split MixtureBase into three abstract classes ObsMixin, HiddenMixin and MixtureBase(ObsMixn, HiddenMixin). I also implemented subclasses
for Gaussian Mixture ObsGaussianMixin(ObsMixin), MixtureMixin(HiddenMixin), GaussianMixture(MixtureBase, ObsGaussianMixin, MixtureMixin), but Python does allow me to do this due to there is correct MRO. :-|. I changed them back, but this
unsuccessful experiment gives me a nice base class, MixtureBase.
I also tried to use cached_property to store the intermediate variables such as, $\ln \pi$, $\ln \Lambda$, and cholsky decomposed $ W^-1 $, but didn’t get much benefits. It is almost the same to save these variables as private attributes into instances.
The numerical issue comes from responsibility is extremely small. When estimating resp * log resp, it gives NAN. I simply avoid computing when resp < 10*EPS. Still, ELBO seems suspicious.
The current implementation of VBGMM in scikit-learn cannot learn the correct parameters on old-faithful data.
VBGMM(alpha=0.0001, covariance_type='full', init_params='wmc',
min_covar=None, n_components=6, n_iter=100, params='wmc',
random_state=None, thresh=None, tol=0.001, verbose=0)
It gives only one components. The weights_ is
array([ 7.31951611e-07, 7.31951611e-07, 7.31951611e-07,
7.31951611e-07, 7.31951611e-07, 9.99996340e-01])
I also implemented DirichletProcessGaussianMixture. But currently it looks the same as BayesianGaussianMixture.
Both of them can infer the best number of components. DirichletProcessGaussianMixture took a slightly more iteration
than BayesianGaussianMixture. If we infer Dirichlet Process Mixture by Gibbs sampling, we don’t need to specify the
truncated level, only alpha the concentration parameter is enough. But with variational inference, we still need
the give the model the maximal possible number of components, i.e., the truncated level $T$.