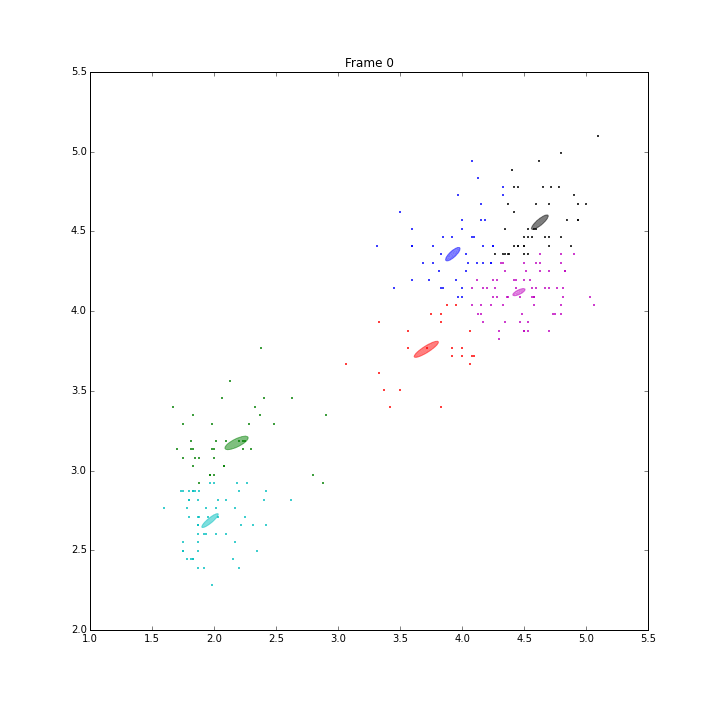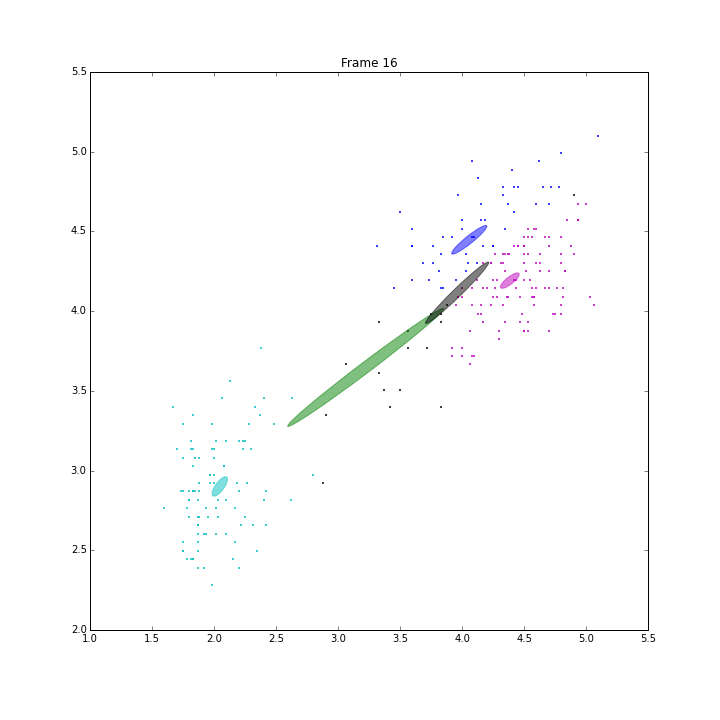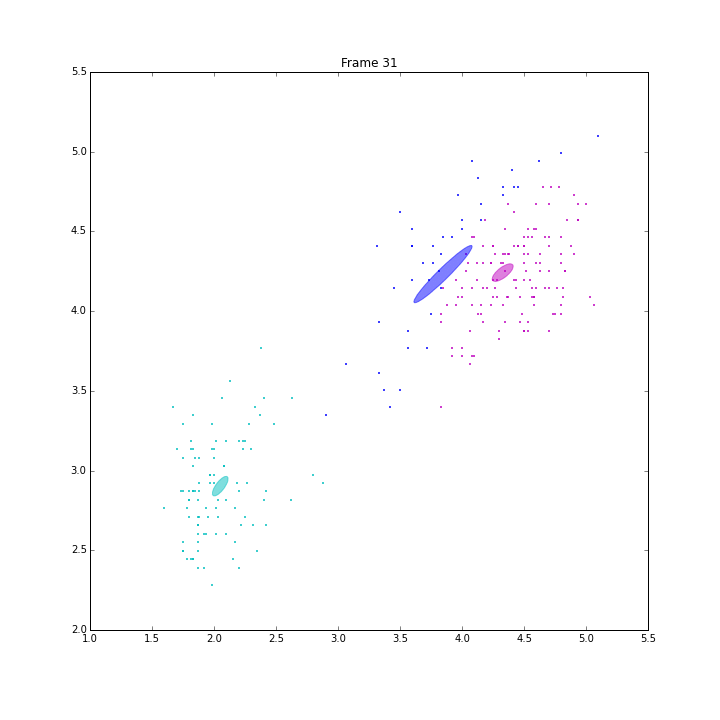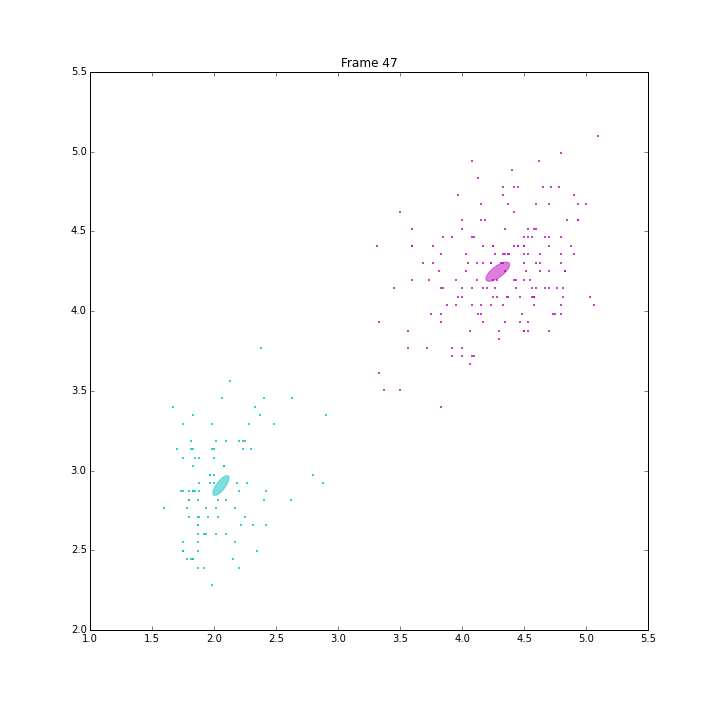
GSoC is approaching its end. I am very glad to have such great experience this summer. I explored the classical machine learning models, Gaussian mixture models (GM), Bayesian Gaussian mixture models with variational inferences (BGM), and Dirichlet Process Gaussian mixture (DPGM). The code and doc is in PR4802.
- Incompatible Interface
- Issue 1528 Consistency in GMM, _get_covars Covariance and precision matrix are interchangeably used in different literature. The
covariancematrices that GMM uses are not provided in VBGMM and DPGMM. In PR4802, GM, BGM, DPGM have common interface, i.e. the propertiesweights_,means_,covariances_, although in BGM and DPGM provide the expected parameters - Issue 2473 Bug: GMM
score()returns an array, not a valueGMM.scorereturns the per-sample likelihood, where as other models return the sum of likelihood. In PR4802,score()is defined as the normalized likelihood, which is a scalar. - Issue 3813 log-responsibilities in GMM
GMM.score_samplesreturns exponentiated log-responsibility which has possibility of precision loss and overflow. In PR4802,score_samples()returns the log of weighted probabilities. - Issue 4062 KernelDensity and GMM interfaces are unnecessarily confusing
- Issue 5129 PCA.score is log density right? 4062, 5129 are both about the concept of
score, currently there is no clear conclusion, PR4802 use the log-likelihood definition - Issue 4429 incorrect estimated means lead to non positive definite covariance in GMM In this issue, I found that GMM does not provide an explicit way for users to initialize
means,covariancesandweights. PR4802 allows user to initialize the parameters of three models in_init__, although there are many initial parameters for the last two models
- Issue 1528 Consistency in GMM, _get_covars Covariance and precision matrix are interchangeably used in different literature. The
- Potential bugs
- Issue 1764 DPGMM - _update_concentrations fail implementation A bug when updating
gamma_. It has been fixed. - Issue 2454 Scaling kills DPGMM DPGMM and VBGMM returns incorrect parameter estimations even on simple toy data set.
- Issue 4267 Density doesn’t normalize in VBGMM and DPGMM The
score_samplesmethod of GMM, VBGMM and DPGMM returns the probability density at given values, however the integration of these values does not equal to 1. 2454 and 4267 might because of the problems in the master implementation, now the integral is sum to 1 - Issue 4429 incorrect estimated means lead to non positive definite covariance in GMM Users might set
paramsthat broke GMM training. 4802 prevents users choosing what parameters should be updated, which is indefinite behavior
- Issue 1764 DPGMM - _update_concentrations fail implementation A bug when updating
- Documentation
- VBGMM DPGMM derivation The derivations in these pages are not consistent with the text book such as PRML[14] and MLAPP[15]. I gave much detailed derivation in PDF. For those who are interested in could find it the PR4802
- Testing
- VBGMM and DPGMM have not been tested comprehensively. Although I could not find a good way to test the implemented equations, I did a comprehensive testing on 1D 1-component data set. The total coverage is more than 95%.
Besides these issues, I did some animations and IPN for these three models.
In conclusion, I finished the tasks of in the proposal, but I didn’t have time to do the optional tasks, i.e., the incremental EM algorithm and different covariance estimators. Anyway, after GSoC, I will continue to contribute to the scikit-learn project.